Dna Transcription & Translation Worksheet

Simplified diagram of mRNA synthesis and processing. Enzymes not shown.
Transcription is the procedure of copying a segment of Dna into RNA. The segments of DNA transcribed into RNA molecules that tin can encode proteins are said to produce messenger RNA (mRNA). Other segments of Deoxyribonucleic acid are copied into RNA molecules called non-coding RNAs (ncRNAs). mRNA comprises only 1-iii% of total RNA samples.[1] Less than 2% of the human genome can be transcribed into mRNA (Human genome#Coding vs. noncoding DNA), while at to the lowest degree eighty% of mammalian genomic DNA can exist actively transcribed (in one or more types of cells), with the majority of this 80% considered to be ncRNA.[2]
Both Dna and RNA are nucleic acids, which use base pairs of nucleotides equally a complementary linguistic communication. During transcription, a DNA sequence is read by an RNA polymerase, which produces a complementary, antiparallel RNA strand called a primary transcript.
Transcription gain in the following general steps:
- RNA polymerase, together with ane or more full general transcription factors, binds to promoter DNA.
- RNA polymerase generates a transcription bubble, which separates the ii strands of the Dna helix. This is done by breaking the hydrogen bonds between complementary DNA nucleotides.
- RNA polymerase adds RNA nucleotides (which are complementary to the nucleotides of ane Deoxyribonucleic acid strand).
- RNA sugar-phosphate backbone forms with assistance from RNA polymerase to course an RNA strand.
- Hydrogen bonds of the RNA–Dna helix suspension, freeing the newly synthesized RNA strand.
- If the cell has a nucleus, the RNA may be farther processed. This may include polyadenylation, capping, and splicing.
- The RNA may remain in the nucleus or go out to the cytoplasm through the nuclear pore complex.
If the stretch of DNA is transcribed into an RNA molecule that encodes a protein, the RNA is termed messenger RNA (mRNA); the mRNA, in plough, serves as a template for the protein'due south synthesis through translation. Other stretches of Dna may be transcribed into pocket-sized non-coding RNAs such equally microRNA, transfer RNA (tRNA), pocket-size nucleolar RNA (snoRNA), small nuclear RNA (snRNA), or enzymatic RNA molecules chosen ribozymes[3] too every bit larger non-coding RNAs such as ribosomal RNA (rRNA), and long not-coding RNA (lncRNA). Overall, RNA helps synthesize, regulate, and process proteins; it therefore plays a fundamental office in performing functions inside a cell.
In virology, the term transcription may too be used when referring to mRNA synthesis from an RNA molecule (i.e., equivalent to RNA replication). For case, the genome of a negative-sense single-stranded RNA (ssRNA -) virus may be a template for a positive-sense single-stranded RNA (ssRNA +)[ clarification needed ]. This is because the positive-sense strand contains the sequence data needed to interpret the viral proteins needed for viral replication. This process is catalyzed by a viral RNA replicase.[4] [ clarification needed ]
Background [edit]
A Dna transcription unit encoding for a poly peptide may contain both a coding sequence, which will exist translated into the protein, and regulatory sequences, which direct and regulate the synthesis of that poly peptide. The regulatory sequence before ("upstream" from) the coding sequence is called the five prime untranslated region (v'UTR); the sequence later on ("downstream" from) the coding sequence is called the three prime untranslated region (three'UTR).[three]
As opposed to DNA replication, transcription results in an RNA complement that includes the nucleotide uracil (U) in all instances where thymine (T) would accept occurred in a Deoxyribonucleic acid complement.
Only 1 of the two DNA strands serve as a template for transcription. The antisense strand of DNA is read by RNA polymerase from the 3' finish to the 5' stop during transcription (three' → v'). The complementary RNA is created in the reverse direction, in the 5' → three' direction, matching the sequence of the sense strand with the exception of switching uracil for thymine. This directionality is considering RNA polymerase tin can only add nucleotides to the 3' cease of the growing mRNA chain. This use of only the 3' → 5' Dna strand eliminates the need for the Okazaki fragments that are seen in Dna replication.[3] This likewise removes the demand for an RNA primer to initiate RNA synthesis, every bit is the case in Deoxyribonucleic acid replication.
The non-template (sense) strand of Dna is called the coding strand, because its sequence is the aforementioned as the newly created RNA transcript (except for the substitution of uracil for thymine). This is the strand that is used by convention when presenting a DNA sequence.[5]
Transcription has some proofreading mechanisms, simply they are fewer and less effective than the controls for copying DNA. Equally a result, transcription has a lower copying fidelity than Dna replication.[6]
Major steps [edit]
Transcription is divided into initiation, promoter escape, elongation, and termination.[7]
Setting up for transcription [edit]
Enhancers, transcription factors, Mediator complex and DNA loops in mammalian transcription [edit]
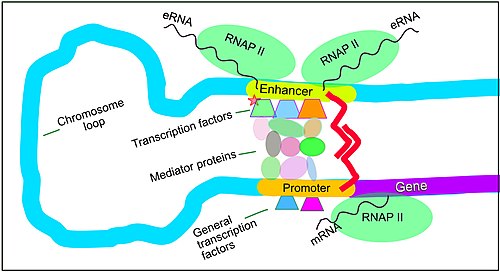
Regulation of transcription in mammals. An active enhancer regulatory region of DNA is enabled to interact with the promoter DNA region of its target cistron past formation of a chromosome loop. This tin initiate messenger RNA (mRNA) synthesis by RNA polymerase II (RNAP 2) bound to the promoter at the transcription start site of the gene. The loop is stabilized by one architectural protein anchored to the enhancer and one anchored to the promoter and these proteins are joined to form a dimer (red zigzags). Specific regulatory transcription factors demark to Deoxyribonucleic acid sequence motifs on the enhancer. Full general transcription factors bind to the promoter. When a transcription factor is activated by a bespeak (hither indicated as phosphorylation shown by a small cerise star on a transcription cistron on the enhancer) the enhancer is activated and tin now activate its target promoter. The active enhancer is transcribed on each strand of Deoxyribonucleic acid in opposite directions past bound RNAP IIs. Mediator (a complex consisting of about 26 proteins in an interacting construction) communicates regulatory signals from the enhancer DNA-bound transcription factors to the promoter.
Setting upward for transcription in mammals is regulated by many cis-regulatory elements, including core promoter and promoter-proximal elements that are located about the transcription starting time sites of genes. Core promoters combined with general transcription factors are sufficient to direct transcription initiation, but generally accept low basal activity.[8] Other important cis-regulatory modules are localized in Deoxyribonucleic acid regions that are afar from the transcription showtime sites. These include enhancers, silencers, insulators and tethering elements.[9] Amidst this constellation of elements, enhancers and their associated transcription factors have a leading part in the initiation of factor transcription.[x] An enhancer localized in a DNA region distant from the promoter of a gene tin have a very large effect on gene transcription, with some genes undergoing up to 100-fold increased transcription due to an activated enhancer.[eleven]
Enhancers are regions of the genome that are major cistron-regulatory elements. Enhancers control cell-blazon-specific gene transcription programs, nearly oft by looping through long distances to come up in concrete proximity with the promoters of their target genes.[12] While there are hundreds of thousands of enhancer Deoxyribonucleic acid regions,[thirteen] for a particular type of tissue only specific enhancers are brought into proximity with the promoters that they regulate. In a study of encephalon cortical neurons, 24,937 loops were found, bringing enhancers to their target promoters.[11] Multiple enhancers, each often at tens or hundred of thousands of nucleotides distant from their target genes, loop to their target gene promoters and can coordinate with each other to control transcription of their common target gene.[12]
The schematic illustration in this department shows an enhancer looping around to come into close physical proximity with the promoter of a target factor. The loop is stabilized by a dimer of a connector protein (due east.g. dimer of CTCF or YY1), with one member of the dimer anchored to its binding motif on the enhancer and the other member anchored to its binding motif on the promoter (represented by the red zigzags in the illustration).[xiv] Several jail cell function specific transcription factors (there are almost 1,600 transcription factors in a human cell[15]) generally bind to specific motifs on an enhancer[16] and a small combination of these enhancer-leap transcription factors, when brought close to a promoter by a DNA loop, govern level of transcription of the target gene. Mediator (a complex normally consisting of almost 26 proteins in an interacting structure) communicates regulatory signals from enhancer DNA-bound transcription factors directly to the RNA polymerase II (politician II) enzyme leap to the promoter.[17]
Enhancers, when agile, are more often than not transcribed from both strands of Dna with RNA polymerases acting in two different directions, producing two enhancer RNAs (eRNAs) every bit illustrated in the Figure.[xviii] An inactive enhancer may exist jump by an inactive transcription factor. Phosphorylation of the transcription factor may activate it and that activated transcription factor may and so activate the enhancer to which it is bound (see minor red star representing phosphorylation of transcription factor bound to enhancer in the illustration).[nineteen] An activated enhancer begins transcription of its RNA before activating transcription of messenger RNA from its target gene.[20]
CpG island methylation and demethylation [edit]

This shows where the methyl group is added when 5-methylcytosine is formed
Transcription regulation at most lx% of promoters is also controlled by methylation of cytosines within CpG dinucleotides (where 5' cytosine is followed past 3' guanine or CpG sites). v-methylcytosine (5-mC) is a methylated form of the DNA base cytosine (see Figure). five-mC is an epigenetic marking found predominantly within CpG sites. About 28 meg CpG dinucleotides occur in the human genome.[21] In most tissues of mammals, on average, 70% to 80% of CpG cytosines are methylated (forming 5-methylCpG or five-mCpG).[22] Methylated cytosines within 5'cytosine-guanine 3' sequences often occur in groups, chosen CpG islands. Nearly threescore% of promoter sequences have a CpG island while simply about half dozen% of enhancer sequences have a CpG island.[23] CpG islands constitute regulatory sequences, since if CpG islands are methylated in the promoter of a gene this tin can reduce or silence gene transcription.[24]
Deoxyribonucleic acid methylation regulates gene transcription through interaction with methyl binding domain (MBD) proteins, such as MeCP2, MBD1 and MBD2. These MBD proteins bind near strongly to highly methylated CpG islands.[25] These MBD proteins have both a methyl-CpG-bounden domain too as a transcription repression domain.[25] They demark to methylated DNA and guide or direct protein complexes with chromatin remodeling and/or histone modifying activity to methylated CpG islands. MBD proteins mostly repress local chromatin such every bit by catalyzing the introduction of repressive histone marks, or creating an overall repressive chromatin environs through nucleosome remodeling and chromatin reorganization.[25]
As noted in the previous department, transcription factors are proteins that bind to specific Deoxyribonucleic acid sequences in order to regulate the expression of a factor. The binding sequence for a transcription factor in DNA is commonly about 10 or 11 nucleotides long. As summarized in 2009, Vaquerizas et al. indicated there are approximately 1,400 different transcription factors encoded in the human genome by genes that constitute virtually 6% of all man protein encoding genes.[26] About 94% of transcription cistron binding sites (TFBSs) that are associated with signal-responsive genes occur in enhancers while only most half-dozen% of such TFBSs occur in promoters.[xvi]
EGR1 protein is a item transcription factor that is of import for regulation of methylation of CpG islands. An EGR1 transcription factor bounden site is ofttimes located in enhancer or promoter sequences.[27] There are almost 12,000 binding sites for EGR1 in the mammalian genome and about half of EGR1 binding sites are located in promoters and half in enhancers.[27] The binding of EGR1 to its target DNA binding site is insensitive to cytosine methylation in the Deoxyribonucleic acid.[27]
While only small amounts of EGR1 transcription factor protein are detectable in cells that are un-stimulated, translation of the EGR1 cistron into protein at i hour afterwards stimulation is drastically elevated.[28] Production of EGR1 transcription factor proteins, in various types of cells, can be stimulated by growth factors, neurotransmitters, hormones, stress and injury.[28] In the encephalon, when neurons are activated, EGR1 proteins are upward-regulated and they bind to (recruit) the pre-existing TET1 enzymes that are produced in high amounts in neurons. TET enzymes can catalyse demethylation of five-methylcytosine. When EGR1 transcription factors bring TET1 enzymes to EGR1 bounden sites in promoters, the TET enzymes can demethylate the methylated CpG islands at those promoters. Upon demethylation, these promoters tin then initiate transcription of their target genes. Hundreds of genes in neurons are differentially expressed after neuron activation through EGR1 recruitment of TET1 to methylated regulatory sequences in their promoters.[27]
The methylation of promoters is besides altered in response to signals. The three mammalian DNA methyltransferasess (DNMT1, DNMT3A, and DNMT3B) catalyze the improver of methyl groups to cytosines in Deoxyribonucleic acid. While DNMT1 is a "maintenance" methyltransferase, DNMT3A and DNMT3B can behave out new methylations. At that place are as well two splice poly peptide isoforms produced from the DNMT3A gene: Deoxyribonucleic acid methyltransferase proteins DNMT3A1 and DNMT3A2.[29]
The splice isoform DNMT3A2 behaves like the production of a classical immediate-early gene and, for example, information technology is robustly and transiently produced afterward neuronal activation.[thirty] Where the Dna methyltransferase isoform DNMT3A2 binds and adds methyl groups to cytosines appears to be determined by histone mail service translational modifications.[31] [32] [33]
On the other hand, neural activation causes degradation of DNMT3A1 accompanied by reduced methylation of at least one evaluated targeted promoter.[34]
Initiation [edit]
Transcription begins with the binding of RNA polymerase, together with one or more full general transcription factors, to a specific DNA sequence referred to as a "promoter" to class an RNA polymerase-promoter "closed complex". In the "closed complex" the promoter DNA is still fully double-stranded.[7]
RNA polymerase, assisted by ane or more full general transcription factors, then unwinds approximately xiv base pairs of DNA to form an RNA polymerase-promoter "open complex". In the "open up complex" the promoter Dna is partly unwound and single-stranded. The exposed, unmarried-stranded DNA is referred to every bit the "transcription bubble."[seven]
RNA polymerase, assisted past i or more general transcription factors, then selects a transcription offset site in the transcription chimera, binds to an initiating NTP and an extending NTP (or a short RNA primer and an extending NTP) complementary to the transcription beginning site sequence, and catalyzes bond germination to yield an initial RNA product.[7]
In bacteria, RNA polymerase holoenzyme consists of v subunits: 2 α subunits, ane β subunit, 1 β' subunit, and 1 ω subunit. In bacteria, there is one general RNA transcription factor known equally a sigma factor. RNA polymerase core enzyme binds to the bacterial general transcription (sigma) factor to form RNA polymerase holoenzyme and then binds to a promoter.[seven] (RNA polymerase is called a holoenzyme when sigma subunit is attached to the core enzyme which is consist of ii α subunits, i β subunit, 1 β' subunit simply). Unlike eukaryotes, the initiating nucleotide of nascent bacterial mRNA is not capped with a modified guanine nucleotide. The initiating nucleotide of bacterial transcripts bears a 5′ triphosphate (v′-PPP), which can exist used for genome-broad mapping of transcription initiation sites.[35]
In archaea and eukaryotes, RNA polymerase contains subunits homologous to each of the five RNA polymerase subunits in bacteria and also contains additional subunits. In archaea and eukaryotes, the functions of the bacterial general transcription factor sigma are performed by multiple general transcription factors that work together.[7] In archaea, there are iii general transcription factors: TBP, TFB, and TFE. In eukaryotes, in RNA polymerase II-dependent transcription, at that place are six general transcription factors: TFIIA, TFIIB (an ortholog of archaeal TFB), TFIID (a multisubunit factor in which the central subunit, TBP, is an ortholog of archaeal TBP), TFIIE (an ortholog of archaeal TFE), TFIIF, and TFIIH. The TFIID is the first component to demark to Deoxyribonucleic acid due to bounden of TBP, while TFIIH is the last component to be recruited. In archaea and eukaryotes, the RNA polymerase-promoter closed complex is usually referred to as the "preinitiation complex."[36]
Transcription initiation is regulated by additional proteins, known as activators and repressors, and, in some cases, associated coactivators or corepressors, which modulate formation and function of the transcription initiation circuitous.[7]
Promoter escape [edit]
Subsequently the first bond is synthesized, the RNA polymerase must escape the promoter. During this time there is a trend to release the RNA transcript and produce truncated transcripts. This is called bootless initiation, and is common for both eukaryotes and prokaryotes.[37] Abortive initiation continues to occur until an RNA product of a threshold length of approximately ten nucleotides is synthesized, at which bespeak promoter escape occurs and a transcription elongation circuitous is formed.
Mechanistically, promoter escape occurs through Deoxyribonucleic acid scrunching, providing the energy needed to break interactions between RNA polymerase holoenzyme and the promoter.[38]
In bacteria, information technology was historically idea that the sigma gene is definitely released later promoter clearance occurs. This theory had been known equally the obligate release model. However, later data showed that upon and following promoter clearance, the sigma cistron is released according to a stochastic model known as the stochastic release model.[39]
In eukaryotes, at an RNA polymerase 2-dependent promoter, upon promoter clearance, TFIIH phosphorylates serine 5 on the carboxy terminal domain of RNA polymerase II, leading to the recruitment of capping enzyme (CE).[40] [41] The exact mechanism of how CE induces promoter clearance in eukaryotes is not yet known.
Elongation [edit]

Simple diagram of transcription elongation
1 strand of the Deoxyribonucleic acid, the template strand (or noncoding strand), is used as a template for RNA synthesis. As transcription gain, RNA polymerase traverses the template strand and uses base pairing complementarity with the Deoxyribonucleic acid template to create an RNA re-create (which elongates during the traversal). Although RNA polymerase traverses the template strand from 3' → 5', the coding (non-template) strand and newly formed RNA can too be used every bit reference points, so transcription can be described as occurring five' → three'. This produces an RNA molecule from 5' → 3', an exact copy of the coding strand (except that thymines are replaced with uracils, and the nucleotides are composed of a ribose (5-carbon) sugar where DNA has deoxyribose (one fewer oxygen cantlet) in its sugar-phosphate backbone).[ commendation needed ]
mRNA transcription can involve multiple RNA polymerases on a single DNA template and multiple rounds of transcription (amplification of particular mRNA), so many mRNA molecules can be chop-chop produced from a single copy of a cistron.[ commendation needed ] The characteristic elongation rates in prokaryotes and eukaryotes are about 10-100 nts/sec.[42] In eukaryotes, still, nucleosomes human action every bit major barriers to transcribing polymerases during transcription elongation.[43] [44] In these organisms, the pausing induced past nucleosomes can be regulated by transcription elongation factors such as TFIIS.[44]
Elongation also involves a proofreading machinery that can supplant incorrectly incorporated bases. In eukaryotes, this may stand for with short pauses during transcription that permit appropriate RNA editing factors to bind. These pauses may exist intrinsic to the RNA polymerase or due to chromatin structure.[ citation needed ]
Termination [edit]
Bacteria use two dissimilar strategies for transcription termination – Rho-independent termination and Rho-dependent termination. In Rho-contained transcription termination, RNA transcription stops when the newly synthesized RNA molecule forms a G-C-rich hairpin loop followed past a run of Us. When the hairpin forms, the mechanical stress breaks the weak rU-dA bonds, now filling the Dna–RNA hybrid. This pulls the poly-U transcript out of the active site of the RNA polymerase, terminating transcription. In the "Rho-dependent" type of termination, a protein factor called "Rho" destabilizes the interaction betwixt the template and the mRNA, thus releasing the newly synthesized mRNA from the elongation complex.[45]
Transcription termination in eukaryotes is less well understood than in bacteria, but involves cleavage of the new transcript followed by template-independent addition of adenines at its new 3' end, in a process called polyadenylation.[46]
Role of RNA polymerase in postal service-transcriptional changes in RNA [edit]
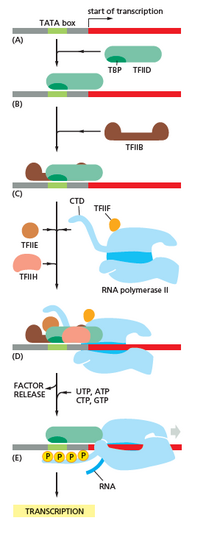
Image showing RNA polymerase interacting with different factors and DNA during transcription, especially CTD (C Terminal Domain)
RNA polymerase plays a very crucial role in all steps including postal service-transcriptional changes in RNA.
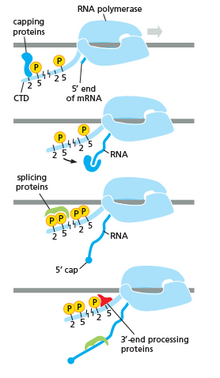
The Image shows how CTD is carrying protein for farther changes in the RNA
Equally shown in the prototype in the right it is axiomatic that the CTD (C Concluding Domain) is a tail that changes its shape; this tail will be used equally a carrier of splicing, capping and polyadenylation, as shown in the paradigm on the left.[47]
Inhibitors [edit]
Transcription inhibitors can be used as antibiotics against, for example, pathogenic bacteria (antibacterials) and fungi (antifungals). An example of such an antibacterial is rifampicin, which inhibits bacterial transcription of DNA into mRNA by inhibiting Dna-dependent RNA polymerase by binding its beta-subunit, while 8-hydroxyquinoline is an antifungal transcription inhibitor.[48] The effects of histone methylation may likewise work to inhibit the action of transcription. Potent, bioactive natural products like triptolide that inhibit mammalian transcription via inhibition of the XPB subunit of the general transcription factor TFIIH has been recently reported as a glucose conjugate for targeting hypoxic cancer cells with increased glucose transporter production.[49]
Endogenous inhibitors [edit]
In vertebrates, the majority of gene promoters incorporate a CpG island with numerous CpG sites.[50] When many of a factor's promoter CpG sites are methylated the gene becomes inhibited (silenced).[51] Colorectal cancers typically take 3 to 6 driver mutations and 33 to 66 hitchhiker or passenger mutations.[52] However, transcriptional inhibition (silencing) may be of more than importance than mutation in causing progression to cancer. For example, in colorectal cancers virtually 600 to 800 genes are transcriptionally inhibited by CpG island methylation (meet regulation of transcription in cancer). Transcriptional repression in cancer tin can also occur past other epigenetic mechanisms, such as altered production of microRNAs.[53] In chest cancer, transcriptional repression of BRCA1 may occur more than frequently by over-produced microRNA-182 than by hypermethylation of the BRCA1 promoter (see Low expression of BRCA1 in breast and ovarian cancers).
Transcription factories [edit]
Active transcription units are clustered in the nucleus, in discrete sites called transcription factories or euchromatin. Such sites can be visualized by allowing engaged polymerases to extend their transcripts in tagged precursors (Br-UTP or Br-U) and immuno-labeling the tagged nascent RNA. Transcription factories can also be localized using fluorescence in situ hybridization or marked by antibodies directed against polymerases. There are ~10,000 factories in the nucleoplasm of a HeLa cell, among which are ~eight,000 polymerase Ii factories and ~2,000 polymerase III factories. Each polymerase Ii factory contains ~8 polymerases. As almost active transcription units are associated with merely one polymerase, each factory usually contains ~8 different transcription units. These units might be associated through promoters and/or enhancers, with loops forming a "cloud" effectually the cistron.[54]
History [edit]
A molecule that allows the genetic textile to be realized equally a poly peptide was get-go hypothesized by François Jacob and Jacques Monod. Severo Ochoa won a Nobel Prize in Physiology or Medicine in 1959 for developing a process for synthesizing RNA in vitro with polynucleotide phosphorylase, which was useful for groovy the genetic code. RNA synthesis by RNA polymerase was established in vitro by several laboratories past 1965; notwithstanding, the RNA synthesized by these enzymes had properties that suggested the beingness of an additional factor needed to terminate transcription correctly.[ citation needed ]
In 1972, Walter Fiers became the starting time person to really testify the existence of the terminating enzyme.
Roger D. Kornberg won the 2006 Nobel Prize in Chemical science "for his studies of the molecular footing of eukaryotic transcription".[55]
Measuring and detecting [edit]
Transcription can be measured and detected in a variety of ways:[ citation needed ]
- G-Less Cassette transcription assay: measures promoter strength
- Run-off transcription analysis: identifies transcription beginning sites (TSS)
- Nuclear run-on assay: measures the relative affluence of newly formed transcripts
- KAS-seq: measures single-stranded Deoxyribonucleic acid generated by RNA polymerases; can work with 1,000 cells.[56]
- RNase protection assay and Chip-Chip of RNAP: observe active transcription sites
- RT-PCR: measures the absolute abundance of total or nuclear RNA levels, which may withal differ from transcription rates
- DNA microarrays: measures the relative affluence of the global total or nuclear RNA levels; notwithstanding, these may differ from transcription rates
- In situ hybridization: detects the presence of a transcript
- MS2 tagging: by incorporating RNA stem loops, such equally MS2, into a factor, these go incorporated into newly synthesized RNA. The stem loops can then exist detected using a fusion of GFP and the MS2 coat protein, which has a high analogousness, sequence-specific interaction with the MS2 stem loops. The recruitment of GFP to the site of transcription is visualized as a single fluorescent spot. This new approach has revealed that transcription occurs in discontinuous bursts, or pulses (see Transcriptional bursting). With the notable exception of in situ techniques, most other methods provide cell population averages, and are not capable of detecting this key property of genes.[57]
- Northern absorb: the traditional method, and until the advent of RNA-Seq, the most quantitative
- RNA-Seq: applies adjacent-generation sequencing techniques to sequence whole transcriptomes, which allows the measurement of relative abundance of RNA, every bit well every bit the detection of additional variations such every bit fusion genes, post-transcriptional edits and novel splice sites
- Single prison cell RNA-Seq: amplifies and reads fractional transcriptomes from isolated cells, allowing for detailed analyses of RNA in tissues, embryos, and cancers
Reverse transcription [edit]
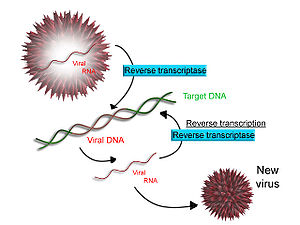
Some viruses (such as HIV, the crusade of AIDS), accept the ability to transcribe RNA into Deoxyribonucleic acid. HIV has an RNA genome that is opposite transcribed into Deoxyribonucleic acid. The resulting Dna can exist merged with the DNA genome of the host cell. The main enzyme responsible for synthesis of DNA from an RNA template is called reverse transcriptase.
In the case of HIV, reverse transcriptase is responsible for synthesizing a complementary Deoxyribonucleic acid strand (cDNA) to the viral RNA genome. The enzyme ribonuclease H so digests the RNA strand, and reverse transcriptase synthesises a complementary strand of Deoxyribonucleic acid to form a double helix DNA structure ("cDNA"). The cDNA is integrated into the host prison cell'south genome by the enzyme integrase, which causes the host cell to generate viral proteins that reassemble into new viral particles. In HIV, subsequent to this, the host jail cell undergoes programmed jail cell death, or apoptosis of T cells.[58] However, in other retroviruses, the host cell remains intact every bit the virus buds out of the cell.
Some eukaryotic cells contain an enzyme with reverse transcription activity chosen telomerase. Telomerase is a reverse transcriptase that lengthens the ends of linear chromosomes. Telomerase carries an RNA template from which it synthesizes a repeating sequence of Deoxyribonucleic acid, or "junk" DNA. This repeated sequence of Deoxyribonucleic acid is called a telomere and can be thought of equally a "cap" for a chromosome. It is important because every time a linear chromosome is duplicated, it is shortened. With this "junk" DNA or "cap" at the ends of chromosomes, the shortening eliminates some of the not-essential, repeated sequence rather than the protein-encoding DNA sequence, that is farther away from the chromosome stop.
Telomerase is often activated in cancer cells to enable cancer cells to duplicate their genomes indefinitely without losing important protein-coding DNA sequence. Activation of telomerase could be office of the process that allows cancer cells to go immortal. The immortalizing factor of cancer via telomere lengthening due to telomerase has been proven to occur in 90% of all carcinogenic tumors in vivo with the remaining 10% using an alternative telomere maintenance route called ALT or Alternative Lengthening of Telomeres.[59]
See also [edit]
- Life
- Cell (biology)
- Prison cell sectionalisation
- DBTSS
- gene
- factor regulation
- gene expression
- Epigenetics
- Genome
- Crick'due south fundamental dogma, in which the product of transcription, mRNA, is translated to form polypeptides, and where information technology is asserted that the contrary processes never occur
- Gene regulation
- Long non-coding RNA
- Missense mRNA
- Splicing - procedure of removing introns from precursor messenger RNA (pre-mRNA) to make messenger RNA (mRNA)
- Transcriptomics
- Translation (biology)
References [edit]
- ^ "RNA Quality and RNA Sample Cess - The states". www.thermofisher.com . Retrieved 2022-08-xxx .
- ^ Li J, Liu C (2019). "Coding or Noncoding, the Converging Concepts of RNAs". Front Genet. 10: 496. doi:x.3389/fgene.2019.00496. PMC6538810. PMID 31178900.
- ^ a b c Eldra P. Solomon, Linda R. Berg, Diana W. Martin. Biology, 8th Edition, International Student Edition. Thomson Brooks/Cole. ISBN 978-0495317142
- ^ Koonin EV, Gorbalenya AE, Chumakov KM (July 1989). "Tentative identification of RNA-dependent RNA polymerases of dsRNA viruses and their relationship to positive strand RNA viral polymerases". FEBS Letters. 252 (1–two): 42–6. doi:10.1016/0014-5793(89)80886-5. PMID 2759231. S2CID 36482110.
- ^ "DNA Strands". world wide web.sci.sdsu.edu. Archived from the original on 27 October 2017. Retrieved 1 May 2018.
- ^ Berg J, Tymoczko JL, Stryer L (2006). Biochemistry (6th ed.). San Francisco: West. H. Freeman. ISBN0-7167-8724-five.
- ^ a b c d e f g Watson JD, Baker TA, Bell SP, Gann AA, Levine M, Losick RM (2013). Molecular Biology of the Gene (7th ed.). Pearson.
- ^ Haberle V, Stark A (Oct 2018). "Eukaryotic core promoters and the functional basis of transcription initiation". Nat Rev Mol Cell Biol. 19 (10): 621–637. doi:ten.1038/s41580-018-0028-8. PMC6205604. PMID 29946135.
- ^ Verheul TC, van Hijfte Fifty, Perenthaler E, Barakat TS (2020). "The Why of YY1: Mechanisms of Transcriptional Regulation by Yin Yang 1". Front Prison cell Dev Biol. 8: 592164. doi:10.3389/fcell.2020.592164. PMC7554316. PMID 33102493.
- ^ Spitz F, Furlong EE (September 2012). "Transcription factors: from enhancer binding to developmental control". Nat Rev Genet. thirteen (9): 613–26. doi:x.1038/nrg3207. PMID 22868264. S2CID 205485256.
- ^ a b Beagan JA, Pastuzyn ED, Fernandez LR, Guo MH, Feng K, Titus KR, Chandrashekar H, Shepherd JD, Phillips-Cremins JE (June 2020). "Three-dimensional genome restructuring beyond timescales of activity-induced neuronal cistron expression". Nat Neurosci. 23 (6): 707–717. doi:ten.1038/s41593-020-0634-6. PMC7558717. PMID 32451484.
- ^ a b Schoenfelder Southward, Fraser P (Baronial 2019). "Long-range enhancer-promoter contacts in gene expression control". Nat Rev Genet. twenty (8): 437–455. doi:10.1038/s41576-019-0128-0. PMID 31086298. S2CID 152283312.
- ^ Pennacchio LA, Bickmore W, Dean A, Nobrega MA, Bejerano G (Apr 2013). "Enhancers: 5 essential questions". Nat Rev Genet. xiv (iv): 288–95. doi:10.1038/nrg3458. PMC4445073. PMID 23503198.
- ^ Weintraub AS, Li CH, Zamudio AV, Sigova AA, Hannett NM, Mean solar day DS, Abraham BJ, Cohen MA, Nabet B, Buckley DL, Guo YE, Hnisz D, Jaenisch R, Bradner JE, Greyness NS, Young RA (Dec 2017). "YY1 Is a Structural Regulator of Enhancer-Promoter Loops". Cell. 171 (7): 1573–1588.e28. doi:10.1016/j.cell.2017.11.008. PMC5785279. PMID 29224777.
- ^ Lambert SA, Jolma A, Campitelli LF, Das PK, Yin Y, Albu M, Chen X, Taipale J, Hughes TR, Weirauch MT (February 2018). "The Man Transcription Factors". Jail cell. 172 (4): 650–665. doi:10.1016/j.jail cell.2018.01.029. PMID 29425488.
- ^ a b Grossman SR, Engreitz J, Ray JP, Nguyen Thursday, Hacohen Due north, Lander ES (July 2018). "Positional specificity of different transcription cistron classes within enhancers". Proc Natl Acad Sci U S A. 115 (30): E7222–E7230. doi:10.1073/pnas.1804663115. PMC6065035. PMID 29987030.
- ^ Allen BL, Taatjes DJ (March 2015). "The Mediator complex: a central integrator of transcription". Nat Rev Mol Cell Biol. 16 (iii): 155–66. doi:10.1038/nrm3951. PMC4963239. PMID 25693131.
- ^ Mikhaylichenko O, Bondarenko V, Harnett D, Schor IE, Males Chiliad, Viales RR, Furlong EE (January 2018). "The degree of enhancer or promoter action is reflected by the levels and directionality of eRNA transcription". Genes Dev. 32 (ane): 42–57. doi:10.1101/gad.308619.117. PMC5828394. PMID 29378788.
- ^ Li QJ, Yang SH, Maeda Y, Sladek FM, Sharrocks AD, Martins-Green Grand (January 2003). "MAP kinase phosphorylation-dependent activation of Elk-ane leads to activation of the co-activator p300". EMBO J. 22 (2): 281–91. doi:10.1093/emboj/cdg028. PMC140103. PMID 12514134.
- ^ Carullo NV, Phillips I RA, Simon RC, Soto SA, Hinds JE, Salisbury AJ, Revanna JS, Bunner KD, Ianov Fifty, Sultan FA, Savell KE, Gersbach CA, Solar day JJ (September 2020). "Enhancer RNAs predict enhancer-factor regulatory links and are critical for enhancer function in neuronal systems". Nucleic Acids Res. 48 (17): 9550–9570. doi:10.1093/nar/gkaa671. PMC7515708. PMID 32810208.
- ^ Lövkvist C, Dodd IB, Sneppen K, Haerter JO (June 2016). "Dna methylation in man epigenomes depends on local topology of CpG sites". Nucleic Acids Res. 44 (11): 5123–32. doi:x.1093/nar/gkw124. PMC4914085. PMID 26932361.
- ^ Jabbari K, Bernardi K (May 2004). "Cytosine methylation and CpG, TpG (CpA) and TpA frequencies". Gene. 333: 143–ix. doi:10.1016/j.gene.2004.02.043. PMID 15177689.
- ^ Steinhaus R, Gonzalez T, Seelow D, Robinson PN (June 2020). "Pervasive and CpG-dependent promoter-similar characteristics of transcribed enhancers". Nucleic Acids Res. 48 (x): 5306–5317. doi:ten.1093/nar/gkaa223. PMC7261191. PMID 32338759.
- ^ Bird A (January 2002). "DNA methylation patterns and epigenetic memory". Genes Dev. 16 (1): six–21. doi:10.1101/gad.947102. PMID 11782440.
- ^ a b c Du Q, Luu PL, Stirzaker C, Clark SJ (2015). "Methyl-CpG-binding domain proteins: readers of the epigenome". Epigenomics. vii (6): 1051–73. doi:10.2217/epi.15.39. PMID 25927341.
- ^ Vaquerizas JM, Kummerfeld SK, Teichmann SA, Luscombe NM (April 2009). "A census of human being transcription factors: function, expression and evolution". Nat. Rev. Genet. 10 (4): 252–63. doi:10.1038/nrg2538. PMID 19274049. S2CID 3207586.
- ^ a b c d Sun Z, Xu X, He J, Murray A, Sun MA, Wei X, Wang 10, McCoig E, Xie E, Jiang X, Li L, Zhu J, Chen J, Morozov A, Pickrell AM, Theus MH, Xie H (August 2019). "EGR1 recruits TET1 to shape the brain methylome during development and upon neuronal action". Nat Commun. 10 (1): 3892. Bibcode:2019NatCo..10.3892S. doi:10.1038/s41467-019-11905-3. PMC6715719. PMID 31467272.
- ^ a b Kubosaki A, Tomaru Y, Tagami Grand, Arner Due east, Miura H, Suzuki T, Suzuki M, Suzuki H, Hayashizaki Y (2009). "Genome-wide investigation of in vivo EGR-ane binding sites in monocytic differentiation". Genome Biol. 10 (4): R41. doi:10.1186/gb-2009-10-four-r41. PMC2688932. PMID 19374776.
- ^ Bayraktar Chiliad, Kreutz MR (Apr 2018). "Neuronal DNA Methyltransferases: Epigenetic Mediators between Synaptic Activity and Cistron Expression?". Neuroscientist. 24 (2): 171–185. doi:ten.1177/1073858417707457. PMC5846851. PMID 28513272.
- ^ Oliveira AM, Hemstedt TJ, Bading H (July 2012). "Rescue of crumbling-associated decline in Dnmt3a2 expression restores cerebral abilities". Nat Neurosci. 15 (8): 1111–3. doi:10.1038/nn.3151. PMID 22751036. S2CID 10590208.
- ^ Dhayalan A, Rajavelu A, Rathert P, Tamas R, Jurkowska RZ, Ragozin S, Jeltsch A (August 2010). "The Dnmt3a PWWP domain reads histone 3 lysine 36 trimethylation and guides Deoxyribonucleic acid methylation". J Biol Chem. 285 (34): 26114–xx. doi:10.1074/jbc.M109.089433. PMC2924014. PMID 20547484.
- ^ Manzo M, Wirz J, Ambrosi C, Villaseñor R, Roschitzki B, Baubec T (Dec 2017). "Isoform-specific localization of DNMT3A regulates DNA methylation allegiance at bivalent CpG islands". EMBO J. 36 (23): 3421–3434. doi:10.15252/embj.201797038. PMC5709737. PMID 29074627.
- ^ Dukatz M, Holzer K, Choudalakis M, Emperle M, Lungu C, Bashtrykov P, Jeltsch A (December 2019). "H3K36me2/3 Binding and Deoxyribonucleic acid Binding of the Deoxyribonucleic acid Methyltransferase DNMT3A PWWP Domain Both Contribute to its Chromatin Interaction". J Mol Biol. 431 (24): 5063–5074. doi:10.1016/j.jmb.2019.09.006. PMID 31634469. S2CID 204832601.
- ^ Bayraktar Chiliad, Yuanxiang P, Confettura AD, Gomes GM, Raza SA, Stork O, Tajima S, Suetake I, Karpova A, Yildirim F, Kreutz MR (November 2020). "Synaptic command of DNA methylation involves activity-dependent deposition of DNMT3A1 in the nucleus". Neuropsychopharmacology. 45 (12): 2120–2130. doi:10.1038/s41386-020-0780-2. PMC7547096. PMID 32726795.
- ^ Boutard, Magali (2016). "Global repositioning of transcription start sites in a plant-fermenting bacterium". Nature Communications. vii: 13783. Bibcode:2016NatCo...713783B. doi:x.1038/ncomms13783. PMC5171806. PMID 27982035.
- ^ Roeder, Robert G. (1991). "The complexities of eukaryotic transcription initiation: regulation of preinitiation complex associates". Trends in Biochemical Sciences. 16 (11): 402–408. doi:10.1016/0968-0004(91)90164-Q. ISSN 0968-0004. PMID 1776168.
- ^ Goldman SR, Ebright RH, Nickels Be (May 2009). "Direct detection of bootless RNA transcripts in vivo". Scientific discipline. 324 (5929): 927–8. Bibcode:2009Sci...324..927G. doi:10.1126/science.1169237. PMC2718712. PMID 19443781.
- ^ Revyakin A, Liu C, Ebright RH, Strick TR (November 2006). "Abortive initiation and productive initiation by RNA polymerase involve Dna scrunching". Scientific discipline. 314 (5802): 1139–43. Bibcode:2006Sci...314.1139R. doi:ten.1126/science.1131398. PMC2754787. PMID 17110577.
- ^ Raffaelle M, Kanin EI, Vogt J, Burgess RR, Ansari AZ (November 2005). "Holoenzyme switching and stochastic release of sigma factors from RNA polymerase in vivo". Molecular Cell. 20 (three): 357–66. doi:10.1016/j.molcel.2005.10.011. PMID 16285918.
- ^ Mandal SS, Chu C, Wada T, Handa H, Shatkin AJ, Reinberg D (May 2004). "Functional interactions of RNA-capping enzyme with factors that positively and negatively regulate promoter escape by RNA polymerase II". Proceedings of the National Academy of Sciences of the United States of America. 101 (20): 7572–seven. Bibcode:2004PNAS..101.7572M. doi:10.1073/pnas.0401493101. PMC419647. PMID 15136722.
- ^ Goodrich JA, Tjian R (April 1994). "Transcription factors IIE and IIH and ATP hydrolysis direct promoter clearance by RNA polymerase II". Cell. 77 (1): 145–56. doi:x.1016/0092-8674(94)90242-9. PMID 8156590. S2CID 24602504.
- ^ Milo, Ron; Philips, Rob. "Cell Biological science past the Numbers: What is faster, transcription or translation?". book.bionumbers.org. Archived from the original on 20 April 2017. Retrieved viii March 2017.
- ^ Hodges C, Bintu L, Lubkowska Fifty, Kashlev 1000, Bustamante C (July 2009). "Nucleosomal fluctuations govern the transcription dynamics of RNA polymerase II". Science. 325 (5940): 626–8. Bibcode:2009Sci...325..626H. doi:x.1126/science.1172926. PMC2775800. PMID 19644123.
- ^ a b Fitz V, Shin J, Ehrlich C, Farnung L, Cramer P, Zaburdaev Five, Grill SW (2016). "Nucleosomal organization affects single-molecule transcription dynamics". Proceedings of the National Academy of Sciences. 113 (45): 12733–12738. doi:10.1073/pnas.1602764113. PMC5111697. PMID 27791062.
- ^ Richardson JP (September 2002). "Rho-dependent termination and ATPases in transcript termination". Biochimica et Biophysica Acta (BBA) - Gene Structure and Expression. 1577 (2): 251–260. doi:10.1016/S0167-4781(02)00456-6. PMID 12213656.
- ^ Lykke-Andersen South, Jensen Thursday (October 2007). "Overlapping pathways dictate termination of RNA polymerase Ii transcription". Biochimie. 89 (10): 1177–82. doi:ten.1016/j.biochi.2007.05.007. PMID 17629387.
- ^ Cramer, P.; Armache, Yard.-J.; Baumli, Southward.; Benkert, S.; Brueckner, F.; Buchen, C.; Damsma, G.E.; Dengl, S.; Geiger, S.R.; Jasiak, A.J.; Jawhari, A. (June 2008). "Structure of Eukaryotic RNA Polymerases". Annual Review of Biophysics. 37 (i): 337–352. doi:10.1146/annurev.biophys.37.032807.130008. ISSN 1936-122X. PMID 18573085.
- ^ http://www.sigmaaldrich.com/US/en/product/sial/h6878 eight-Hydroxyquinoline from SIGMA-ALDRICH. Retrieved 2022-02-15
- ^ Datan E, Minn I, Peng Ten, He QL, Ahn H, Yu B, Pomper MG, Liu JO (2020). "A Glucose-Triptolide Conjugate Selectively Targets Cancer Cells nether Hypoxia". iScience. 23 (9): 101536. Bibcode:2020iSci...23j1536D. doi:10.1016/j.isci.2020.101536. PMC7509213. PMID 33083765.
- ^ Saxonov Southward, Berg P, Brutlag DL (January 2006). "A genome-broad analysis of CpG dinucleotides in the human genome distinguishes two singled-out classes of promoters". Proceedings of the National Academy of Sciences of the Usa of America. 103 (5): 1412–7. Bibcode:2006PNAS..103.1412S. doi:x.1073/pnas.0510310103. PMC1345710. PMID 16432200.
- ^ Bird A (January 2002). "Deoxyribonucleic acid methylation patterns and epigenetic memory". Genes & Development. xvi (1): half dozen–21. doi:ten.1101/gad.947102. PMID 11782440.
- ^ Vogelstein B, Papadopoulos N, Velculescu VE, Zhou Southward, Diaz LA, Kinzler KW (March 2013). "Cancer genome landscapes". Science. 339 (6127): 1546–58. Bibcode:2013Sci...339.1546V. doi:10.1126/science.1235122. PMC3749880. PMID 23539594.
- ^ Tessitore A, Cicciarelli One thousand, Del Vecchio F, Gaggiano A, Verzella D, Fischietti M, Vecchiotti D, Capece D, Zazzeroni F, Alesse E (2014). "MicroRNAs in the Dna Harm/Repair Network and Cancer". International Journal of Genomics. 2014: 820248. doi:10.1155/2014/820248. PMC3926391. PMID 24616890.
- ^ Papantonis A, Kohro T, Baboo Due south, Larkin JD, Deng B, Brusk P, Tsutsumi Due south, Taylor S, Kanki Y, Kobayashi M, Li Thou, Poh HM, Ruan X, Aburatani H, Ruan Y, Kodama T, Wada Y, Cook PR (November 2012). "TNFα signals through specialized factories where responsive coding and miRNA genes are transcribed". The EMBO Journal. 31 (23): 4404–14. CiteSeerX10.1.1.919.1919. doi:10.1038/emboj.2012.288. PMC3512387. PMID 23103767.
- ^ "Chemistry 2006". Nobel Foundation. Archived from the original on March 15, 2007. Retrieved March 29, 2007.
- ^ Wu, T (April 2020). "Kethoxal-assisted unmarried-stranded Dna sequencing captures global transcription dynamics and enhancer activeness in situ". Nature Methods. 17 (5): 515–523. doi:10.1038/s41592-020-0797-ix. PMC7205578. PMID 32251394. S2CID 214810294.
- ^ Raj A, van Oudenaarden A (October 2008). "Nature, nurture, or adventure: stochastic gene expression and its consequences". Jail cell. 135 (2): 216–26. doi:10.1016/j.cell.2008.09.050. PMC3118044. PMID 18957198.
- ^ Kolesnikova IN (2000). "Some patterns of apoptosis mechanism during HIV-infection". Dissertation (in Russian). Archived from the original on July 10, 2011. Retrieved February 20, 2011.
- ^ Cesare AJ, Reddel RR (May 2010). "Alternative lengthening of telomeres: models, mechanisms and implications". Nature Reviews Genetics. 11 (5): 319–30. doi:10.1038/nrg2763. PMID 20351727. S2CID 19224032.
External links [edit]
- Interactive Java simulation of transcription initiation. Archived 2011-07-22 at the Wayback Automobile From Center for Models of Life at the Niels Bohr Institute.
- Interactive Coffee simulation of transcription interference--a game of promoter authority in bacterial virus. Archived 2011-08-26 at the Wayback Automobile From Heart for Models of Life at the Niels Bohr Institute.
- Virtual Cell Animation Drove, Introducing Transcription
Dna Transcription & Translation Worksheet,
Source: https://en.wikipedia.org/wiki/Transcription_(biology)
Posted by: broomfife1996.blogspot.com


0 Response to "Dna Transcription & Translation Worksheet"
Post a Comment